Genomic and Transcriptomic Tools
Whole Genome Sequencing (WGS)
Enables identification of genetic variations, resistance markers, and evolutionary relationships among strains.
Provides insight into transcriptional responses of mycobacteria under different environmental conditions.
Comparative Genomics
Supports the discovery of genes linked to virulence and pathogenicity.
Proteomics and Structural Biology
- Mass Spectrometry (MS): Used for large-scale identification of proteins and their modifications.
- Protein Structure Analysis: X-ray crystallography and cryo-electron microscopy reveal structural details of mycobacterial proteins.
- Interactomics: Studies protein–protein interactions involved in survival and immune evasion.
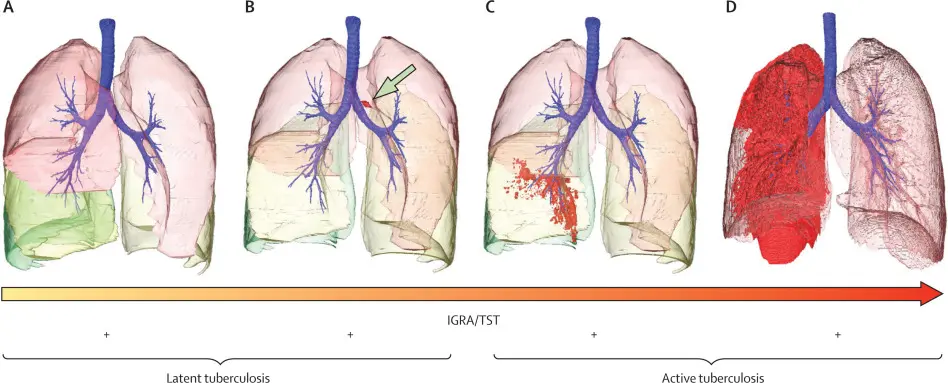
Cell-Based Models
1.Macrophage Infection Assays:
Model the interaction between mycobacteria and host immune cells.
2.Organoid and Tissue Models
Provide advanced systems to mimic lung environments.
3.Fluorescent Reporter Strains
Allow visualization of bacterial growth and stress responses inside host cells.
Bioinformatics and Data Integration
Databases:
Genomic and proteomic databases collect and organize TB-related data.
Systems Biology:
Combines multi-omics datasets to model regulatory networks.
Machine Learning Approaches:
Support prediction of resistance patterns and biomarkers.
Laboratory Techniques
Identifies bacterial genes essential for survival in different conditions.
Measures host cell responses during infection.
Visualizes bacterial localization in host cells and tissues.